Applicants are invited for several Internship positions to undertake research and development for innovative projects. All projects will focus on the implementation of state-of-the-art AI techniques in healthcare applications. Please send your CV and a brief note expressing your interest to the corresponding supervisor.
Please only reach out if you are confident in programming and machine learning. Interviews will include practical programming tasks.
Open Positions
User-Friendly Annotation Tool for Medical Imaging Datasets
Supervisor: Dr Ali Gheitasy
This project aims to develop an accessible and efficient tool for tagging and labeling medical images. The primary objective is to simplify the annotation process using artificial intelligence and user-centered design principles. This tool will be designed to support clinicians, researchers, and students in managing complex imaging datasets by automating repetitive tasks and reducing manual errors.
Key deliverables include a working prototype, user guide, and a peer-reviewed publication. External collaborators include hospitals and med-tech companies providing real-world data and testing environments.
AI-Powered User Interface for Real-time Medical Image Acquisition and Quality Feedback
Supervisor: Dr Nasim Dadashi Serej
This project addresses the need for improved quality control in medical image acquisition. It will develop a real-time feedback interface powered by AI to assist clinicians during image capture. The system will guide users to obtain optimal quality images, reducing the frequency of retakes and improving diagnostic accuracy.
The internship includes developing UI components, integrating AI algorithms, and evaluating the system in clinical settings. Outcomes include a tested prototype and clinical feedback reports.
Feature Selection for Predicting Patenit Outcomes
Supervisor: Prof. Massoud Zolgharni
This project focuses on isolating key clinical and imaging features from large datasets. These features will serve as inputs for predictive models of patient outcomes, improving the precision and interpretability of prognosis tools.
The intern will apply feature selection techniques like Lasso and correlation analysis and validate findings with clinical partners. Deliverables include a validated feature set, collaborative publications, and knowledge transfer to students involved in healthcare data science.
Closed Positions
Role 1: Automated Early Detection of Barrett’s Esophagus (BE) in Endoscopic Images
Barrett's Esophagus (BE) is a condition where the normal lining of the esophagus is replaced by
abnormal tissue, increasing the risk of esophageal cancer.
The early detection of BE is crucial
for timely treatment and improved patient outcomes. However, the current method of diagnosing
BE is through endoscopy, which is an invasive and time-consuming procedure.
Moreover, the visual assessment of endoscopic images for BE detection is
subjective and prone to inter-observer variability.
To overcome these limitations, there is a need for an automated system that can detect BE in
endoscopic images accurately and efficiently. The aim of this project is to develop an
automated early detection system for BE in endoscopic images using machine learning algorithms.
The project will involve the following steps:
- Data Preprocessing to remove noise and artifacts, and to enhance the contrast and sharpness of the images.
This step will improve the quality of the images and ensure better detection accuracy.
- Develop a deep learning Model for automatic classification of the endoscopic images as BE or non-BE.
- Evaluation the performance of the developed model on test set of endoscopic images using evaluation metrics.
- Deploy the developed model as an automated system for early detection of BE in endoscopic images.
The ultimate goal of the project is to provide a cost-effective, non-invasive and reliable screening tool for the early detection of BE, enabling prompt treatment and improving patient outcomes.
This system will help improve the diagnosis of BE and reduce the burden on endoscopists.
Download the document
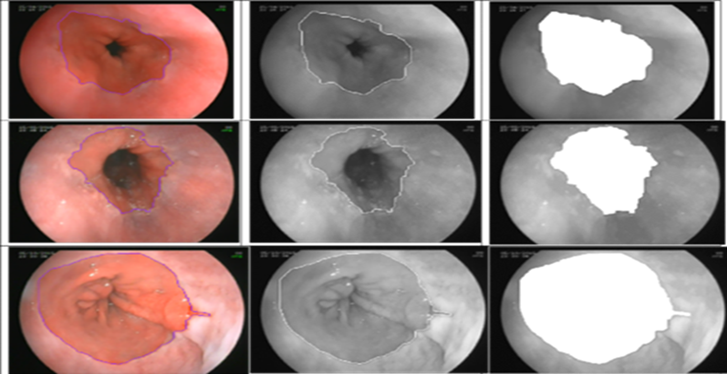
Role 2: Automated Polyp detection and segmentation in Colonoscopy Images
Colorectal cancer is the third most common cancer worldwide, with about 1.8 million new cases and 900,000 deaths per year. Screening for colorectal cancer is essential for early detection and prevention, and colonoscopy is the most effective screening method. However, the effectiveness of colonoscopy depends on the ability of the endoscopist to detect and remove polyps, which are the precursor lesions to colorectal cancer. Missed polyps can lead to delayed diagnosis and treatment, and therefore, improving polyp detection rates is crucial for reducing the incidence and mortality of colorectal cancer.
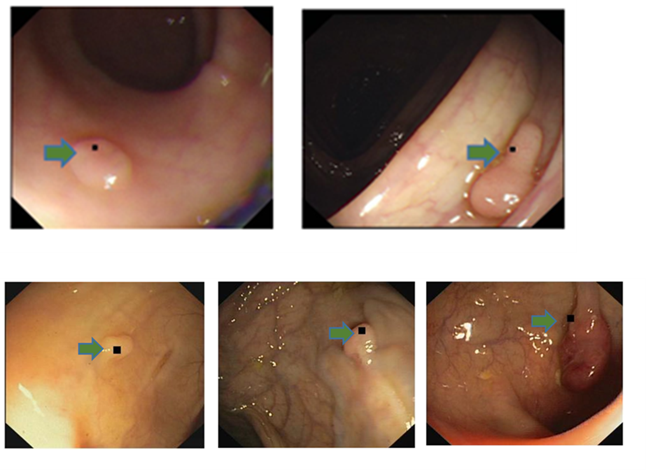
The objective of this project is to improve polyp detection rates in colonoscopy and,
consequently, improve the screening and diagnosis of colorectal cancer.
The project aims to achieve the following objectives:
- Develop a deep learning algorithm for automatic polyp detection and classification in
colonoscopy images.
- Train and validate the algorithm using a large dataset of colonoscopy images with polyps of different sizes, shapes,
and locations.
- Integrate the algorithm into the colonoscopy system to assist endoscopists in
real-time during the procedure.
- Evaluate the performance of the algorithm in terms of sensitivity, specificity, and positive predictive value,
and compare it with the performance of human endoscopists.
- Assess the impact of the algorithm on polyp detection rates, adenoma detection rates, and interval cancer rates in a clinical setting.
Download the document
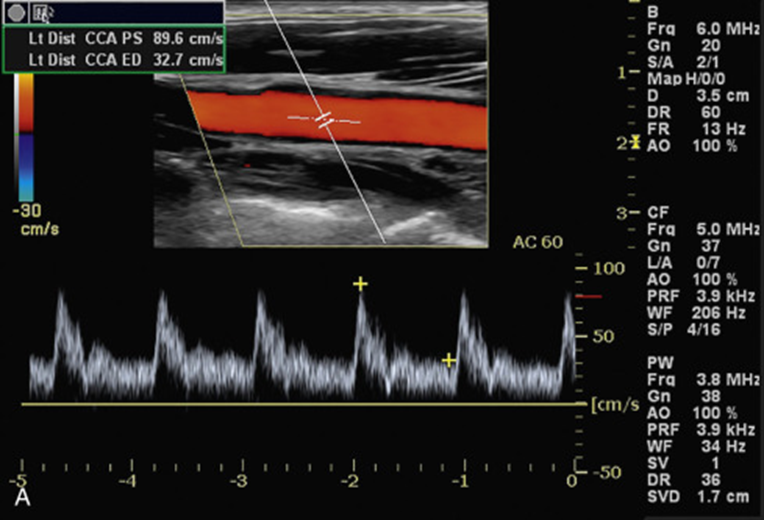
Role 3: Doppler Mitral inflow echocardiographic_ pixel to velocity
This project focuses on the development of a deployable model for converting pixel values to velocity values in Doppler Mitral inflow echocardiographic images. Currently, AI models can detect and measure necessary parameters on Mitral echocardiographic images, but the measurements are acquired in pixel values. In order to adapt these techniques to real-world scenarios, automated measurements should be converted to velocity values.
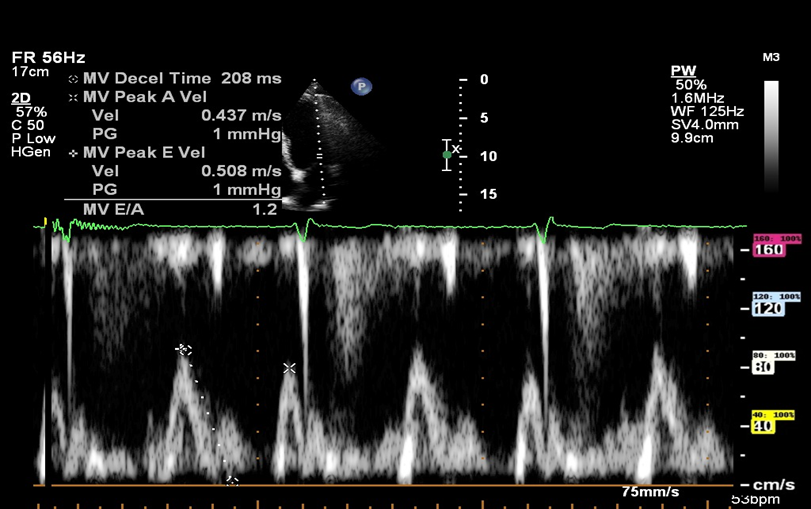
The objective of this project is to develop a deployable model that can process
echocardiographic images and determine the pixel-to-velocity conversion rate. The model will
be developed using computer vision techniques and will be incorporated into a larger deep
learning pipeline.
Expected outcomes: The deployable model developed in this project will enable automated
pixel-to-velocity conversion in Doppler Mitral inflow echocardiographic images.
This will facilitate the translation of AI models for functional assessment of the
Mitral valve into real-world scenarios. The project will contribute to the advancement
of non-invasive cardiac diagnostics, leading to better patient care and outcomes.
Download the document
Role 4: Doppler Flow Imaging and Analysis
Doppler echocardiography is a widely used technique in the functional evaluation of heart valves. However, the current measurement process is laborious and subjective, leading to high intra- and inter-observer variability. The use of automated systems to analyse Doppler echocardiographic images could standardise the measurement process and potentially reduce the time spent on such measurements, leading to improved clinical workflow.